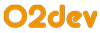33
How Roche Discovered Novel Potential Gene Targets with TypeDB
Debrief from a Vaticle Community talk — featuring David Dylus, Scientist, Systems Biology, at Roche. This talk was delivered virtually at Orbit 2021 in April.
Central to drug discovery is the search for targets that are important in disease mechanisms. However, currently, all known targets have been slowly tried and tested. In this project, David and his team designed a rule system to infer and find hidden connections between targets and diseases.

In the story to follow, David presents how his team at Roche was able to identify potential novel targets that were not identified by Open Targets as highly ranked. This was made possible with TypeDB, which his team used to store the relevant data and then find underlying biological evidence for those new targets.
For this project, three datasets were used: STRING, Oma, and DisGeNET.

Finally, DisGeNET was used to provide variant information, which allowed them to link mutations to genes: for example, if you have a mutation on your genome, if that mutation is linked to a gene, and if that gene is linked to a disease, we can association the disease to that mutation. This database was also mentioned by Tomás in his Orbit 2021 talk [Computational Future of Biology].
This then leads us to ask a question such as:
Do people that have this mutation, also have some type of prevalence for a specific disease?
To answer this, we should check how closely the specific mutation is to the gene and then assign that this variant somehow modulates that gene. This would explain why we see this type of disease phenotype.
To start, the team looked at existing targets on the Open Targets database and selected those already ranked highly and known to have a high association score— due to internal Roche IP, unfortunately, they cannot mention which ones specifically; they have been renamed for understanding.

David called the highly ranked targets taken from Open Targets
billionDollarTarget and bestTarget, which overall have a high association score, in other words, they are strongly linked to the disease that David's team is interested in. Below we’ll see how TypeDB can be used to find targets that don’t rank highly on Open Targets, but are still indirectly modulating the disease and are therefore potentially high-value targets to explore. For this purpose, David built out a set of rules and schema in TypeQL. Below is just a very small excerpt of how such data can be modelled — taken from BioGrakn-Covid, a Vaticle community-led project by Konrad Mysliwiec (Data Science Software Engineer, Roche). Note that this is a selected schema; the full schema can be found within the BioGrakn-Covid schema file.
define
gene sub fully-formed-anatomical-structure,
owns gene-symbol,
owns gene-name,
plays gene-disease-association:associated-gene;
disease sub pathological-function,
owns disease-name,
owns disease-id,
owns disease-type,
plays gene-disease-association:associated-disease;
protein sub chemical,
owns uniprot-id,
owns uniprot-entry-name,
owns uniprot-symbol,
owns uniprot-name,
owns ensembl-protein-stable-id,
owns function-description,
plays protein-disease-association:associated-protein;
protein-disease-association sub relation,
relates associated-protein,
relates associated-disease;
gene-disease-association sub relation,
owns disgenet-score,
relates associated-gene,
relates associated-disease;With the right schema, rules and data inserted, we can write the first query. The relation below is one that David’s team called
gene-disease-inference, with an attribute order:1 to denote that it’s a direct relation. The query looks like this:match
$d isa disease, has disease-name "Disease";
$r ($gene, $d) isa gene-disease-inference, has order 1;
get $r, $d, $gene;The result is below, we can see that the
billionDollarTarget, the bestTarget, and the youWillNeverGuessTarget are linked to Disease. We also see that these three targets are of order: 1, which indicates a direct and previously known association between the disease and the genes. However, the goal is to find novel targets. 
To do this, they write the query shown below. This looks for diseases and genes connected through a
gene-disease-inference relation with order: 2, but explicitly excludes those that already are connected with a gene-disease-inference relation with order:1:match
$d isa disease, has disease-name "Disease";
$r ($gene, $d) isa gene-disease-inference, has order 2;
not {($gene, $d) isa gene-disease-inference, has order 1;};
get $r, $d, $gene;This query returns a completely different list of genes:
whatCouldIBeTarget, awesomeTarget, and thatTarget. All these are targets connected to Disease through a gene-disease-inference with order:2.
If you are not familiar with TypeDB Workbase, you can right-click one of the inferred relations and select “Explain” in the dropdown. This will explain those inferences and tell you how these targets are connected to our disease via typed roles, played by the targets.
If we explain the inferred relation that links
deadTarget, we see that this target is part of the same gene family as youWillNeverGuessTarget with order: 1. This inference was made possible through a rule, which allows us to infer new data based on existing data. In this case, we found a previously unknown indirect interaction between two targets.
The logic behind the rule that gives this inference breaks down as follows:
When:
Then:
For the other novel targets,
awesomeTarget and bestTarget, we see that those inferences are based on a protein-to-protein interaction, which connects to the whatCouldIBeTarget. If we explain that relation, we see that it is connected to the billionDollarTarget via a gene-disease-association, potentially sharing the same variant to the disease. 
Even though
awesomeTarget and thatTarget appear in the Open Targets database for the disease of interest, they ranked very low. That means they had some link to that disease, but not a strong one. TypeDB uncovered new evidence that suggests those targets could be higher ranked.This is how David’s team at Roche was able to leverage TypeDB’s reasoning engine to find novel targets that might have been missed using standard approaches or more direct approaches.

That said, biology is a very complicated field that is constantly evolving. Data sets that have been true in the past might not be true today. We are constantly dealing with new confounders, different methodologies, the noise that is inherent in biology. The goal is to find novel ways of modulating a disease with strong biological evidence that will work.
Having found a novel target does not necessarily mean that this is now a solution or ready for trials. However, it is a great hypothesis to start digging into its efficacy to modulate a specific disease, whether to find a cure or provide better therapeutics to a patient.
Instead of targeting a single protein, more advanced targeting can be done by integrating additional information such as protein complexes and pathways. For example, we could look for several genes that are part of the same pathway. If a drug cannot modulate a single target enough to impose a positive change in the state of a patient, then we might consider targeting multiple points on the same pathway.
David also mentioned he considered extending the rules to, for example, find higher-order relations, to enable the examination of third, fourth or fifth order connections to the specific disease. There is also room for expanding beyond protein-protein interactions and incorporate very specific query constraints. For instance, we could filter that we want genes X and Y to be part of the same pathway, expressed in the same cell type, shown to be up or down-regulated in disease expression, etc. In this way, like boundary conditions, we can increase our target prioritization and make this highly valuable to our process.
A special thank you to David for his work, contribution to the community and for always bringing joy into his work.
All slides used with permission. You can find the full presentation on the Vaticle YouTube channel here.
33